What Happened
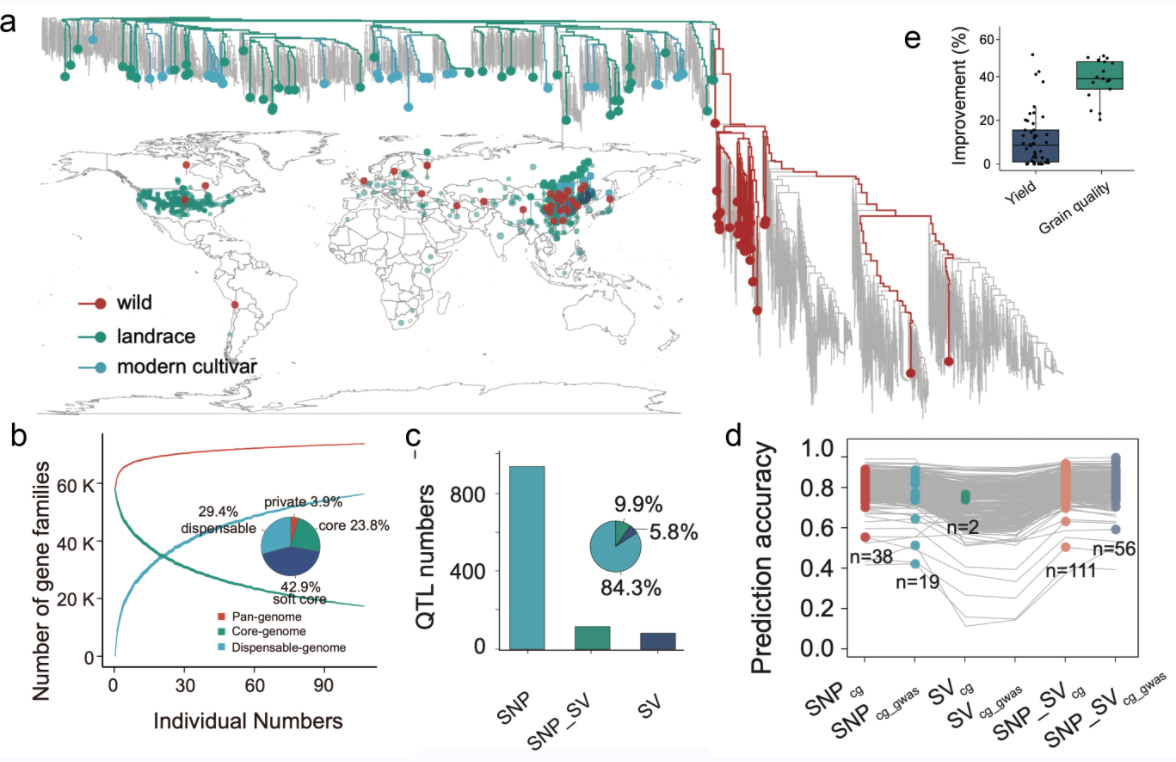
Large-scale genomic and phenomic analyses of modern cultivars empower future rice breeding design
Published: 13 March 2025The Team for the Discovery and Innovative Utilization of Superior Rice Germplasm Resources from the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, in collaboration with the teams for the Safe Preservation and Informatization of Crop Germplasm Resources and Crop Bioinformatics and Big Data, published a paper titled "Large-scale genomic and phenomic" in Molecular Plant The research paper "analyses of modern cultivars empower future rice breeding design". This study conducted deep whole-genome resequencing on 6,044 modern rice varieties from the five major rice-growing areas in China. Combined with 212 sets of phenotypic data from 19 identification points in the five major rice-growing areas, it systematically clarified the genomic and phenotypic selection characteristics of rice varieties in different rice-growing areas, and multi-dimensional revealed the adaptability of varieties in different rice-growing areas and the influence of human selection. Based on the above data, 3,131 QTLS significantly related to key agronomic traits such as panicle stage, yield and stress response were identified, and a new gene OsGL3.6 for regulating rice grain shape was cloned. Further combined with the published data, completed nearly thousands of varieties of rice resources/high density variation of chromatogram, auxiliary tools and develop online breeding, has created the world's largest rice breeding resources integrated data platform - RiceAtlas (https://www.cgris.net/RiceAtlas), Researchers successfully and rapidly improved the grain shape of "Suijing No. 4" rice using this platform. This research achievement provides important theoretical basis and data support for intelligent decision-making in precise design breeding of rice, marking another significant leap in global rice germplasm resources and breeding research following the "3K Rice Genome Project". Original Article Link: https://doi.org/10.1016/j.molp.2025.03.007

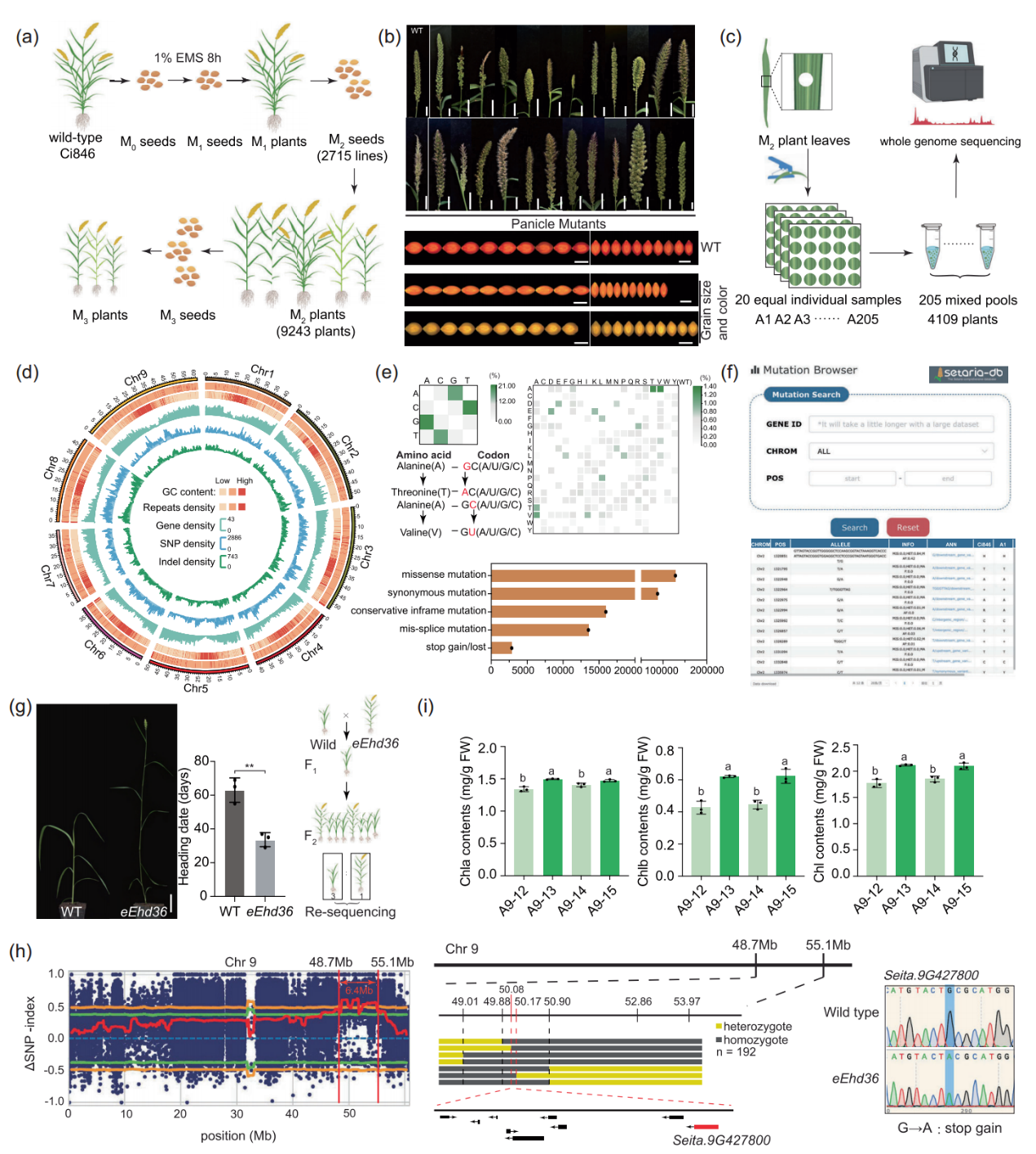
An efficient target-mutant screening platform of model variety Ci846 facilitates genetic studies of Setaria
Published: 03 March 2025On March 3, the team of excellent germplasm resources Exploration and innovative Utilization of characteristic crops of the Institute of Crop Science, Chinese Academy of Agricultural Sciences constructed the genome saturated mutant library and directed mutation screening network database of millet model variety Ci846, which provided efficient research tools for millet functional genome research and molecular design breeding.The relevant research results were published online in the "Plant Biotechnology Journal". Original Article Link: https://onlinelibrary.wiley.com/doi/10.1111/pbi.14594
A High-Quality Foxtail Millet Genome Unlocks a New Chapter for Large-Scale Germplasm Resource Exploration and Utilization
Published: June 9, 2023On June 8, the Research Team for the Exploration and Innovative Utilization of Superior Germplasm Resources of Specialty Crops at the Institute of Crop Sciences, Chinese Academy of Agricultural Sciences, assembled the first high-quality genome map of foxtail millet through genome variation and population structure analyses of foxtail millet germplasm resources. The study systematically elucidated the origin and domestication of foxtail millet, providing population genetics evidence supporting the single-origin hypothesis that "foxtail millet originated in China." Additionally, the team conducted in-depth analyses of 1,844 core germplasm resources of foxtail millet and its wild relative, green foxtail, identifying 1,084 loci and genes associated with important traits. This led to the development of precision and efficient breeding methods based on the genome map of foxtail millet, providing a theoretical foundation and technical pathway for breeding breakthrough varieties. The findings were published in Nature Genetics. Original Article Link: https://www.nature.com/articles/s41588-023-01423-w